- Home >
- Faculty Websites >
- K. Al Nasr
- > Research
Research
Research Interests
|
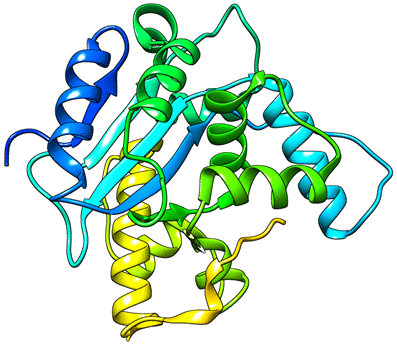
My research interest is centered on developing efficient computational methods for protein structure prediction in de novo modeling. The objective is to provide insights into the function of proteins in nature. Specifically, my research focuses on using Electron Cryo-Microscopy, high performance computing, and graph theory to design algorithms which efficiently predict the structure of proteins in 3-dimensional space.
|
|
Grants
- co-PI, "CREST HBCU-RISE: Advancing Theoretical Artificial Intelligence Infrastructure for Modern Data Science Challenges.", NSF, $1,200,000, 09/15/2024 to 08/31/2027.
- PI, "Excellence in Research: De Novo Protein Modeling, Beyond Topology Determination Problem.", NSF, $535,066, 09/01/2022 - 08/31/2025.
- co-PI, "Strengthen Plant Biotech Program by Integrating Genome Editing and Artificial Intelligence Technology in Tomato Projects.", NIFA, $500,000, 05/01/2022 - 04/30/2025.
- PI, "Data analytics Using Cryo-electron Microscopy Big Data", TSU seed grant, 05/01/2023 to 08/31/2023.
- PI, "Research Initiation Award: Leveraging Protein Structure Determination - An Effective Algorithmic Approach", NSF, $300,000, 04/01/2016 to 03/31/2019.
- PI, "A Computational Tool for Modeling of Protein Macromolecules Guided by Cryo-Electron Microscopy Data", NIH, $399,245, 9/30/2017 to 8/31/2020.
webpage contact:
K. al Nasr


